Methods for calculating solvent accessible surface area (SASA)
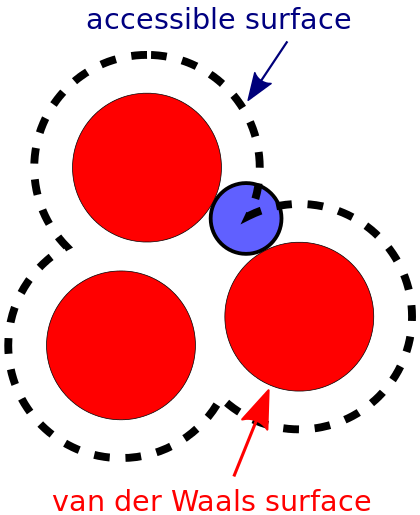
Most of these methods are based on the Shrake-Rupley algorithm [1] which creates many points on the surface of each (heavy) atom and determines whether each point is occluded or exposed to solvent.
You can calculate SASA in VMD’s tkConsole with the measure command using the following syntax:
measure sasa $selection [-points varname] [-restrict restrictedsel] [-samples numsamples]
Other free software packages:
-
Barry Honig SURFace: http://wiki.c2b2.columbia.edu/honiglab_public/index.php/Software:SURFace_Algorithms
-
Python-based ASA by Bosco Ho: http://boscoh.com/protein/calculating-the-solvent-accessible-surface-area-asa.html
-
Calc-surface by Mark Gerstein: download and compile the libproteingeometry package from http://www.molmovdb.org/geometry/
-
GETAREA, a web server: http://curie.utmb.edu/getarea.html
References: [1] A. Shrake, J. A. Rupley, Environment and exposure to solvent of protein atoms. Lysozyme and insulin, Journal of Molecular Biology, Volume 79, Issue 2, 15 September 1973, Pages 351-364, ISSN 0022-2836, DOI: 10.1016/0022-2836(73)90011-9