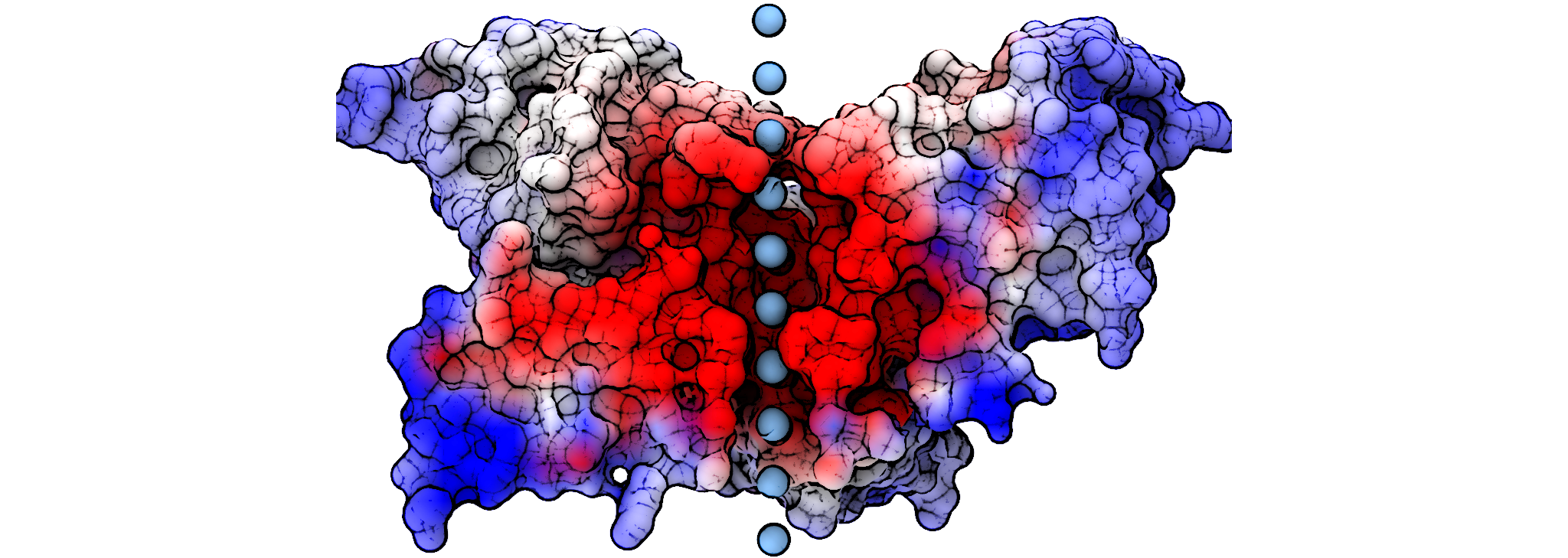
Calculating the ion solvation energy along the permeation pathway of an ion channel
In this tutorial we will model the membrane as a low dielectric slab to calculate the ion solvation permeation pathway. Although this functionality is now built into our APBSmem interface, we will use a script with our draw_membrane Java tool so that we can operate from the command-line.
For more details and the theory behind these calculations, please see my paper in PLoS ONE: http://www.plosone.org/article/info:doi/10.1371/journal.pone.0012722
Requirements
- APBS: poissonboltzmann.org
- Linux, OS X, or Windows with Cygwin
- Java v1.4.2 or greater
Procedure
-
Download and extract the following archive to a new directory: permeation.zip
-
Verify that the tools work for the test case on KcsA by executing this command in a terminal:
./ion_permeation -1.000 1 3
This will move an ion through the pore from -1 to +3 in steps of 1 angstrom.
-
Prepare your protein by converting it to the PQR format. You can use the PDB2PQR server: http://kryptonite.nbcr.net/pdb2pqr/
-
The protein should also be oriented with its principal axis oriented along the z-axis. This can be achieved in the lastest version of APBSmem This can also be done in VMD: http://www.ks.uiuc.edu/Research/vmd/script_library/scripts/orient/ PyMOL: http://www.pymolwiki.org/index.php/Orient or using MODELLER: http://salilab.org/modeller/9v6/manual/node180.html
-
Open the
ion_permeationscript in a text editor. At the top of the script are a series of parameters that specify the membrane geometry and dielectrics. For more details on what these parameters signify, please see our PLoS ONE paper. Change the line that readscat K1K2K3_protein.pqr ion.pqr > K1K2K3_protein.pqr.withion.pqrto reflect your own protein pqr file names. -
Open the permeation.dummy.in and permeation.solv.in input files in a text editor. First make sure the file locations reflect the locations of your pqr files. Now modify the settings to reflect your system. This can be made easier using APBSmem. Details on the parameters are available here: http://www.poissonboltzmann.org/apbs/user-guide/running-apbs/input-files
-
Run the script using the following syntax:
./ion_permeation -1.000 1 3
The arguments are: Starting Z position, Step distance, Stop Z position Please note, the number format of the first argument needs to be 6 characters for the pqr columns to line up properly. -
Results will be saved in results.out.